Projects
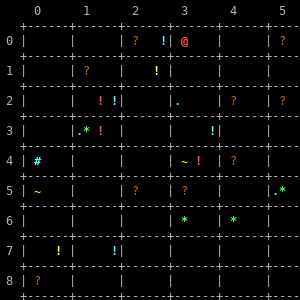
Gal4Xy (personal project)
A turn-based, 4X strategy game, for the moment console-based, written in pure C. The aim is to defeat the AI player(s) and conquer the galaxy. In the near future, Gal4Xy will be multiplayer, meaning you will be able to set up a Gal4Xy server and play it with (or against!) your friends, no LAN required. I just need to figure out how to properly handle data serialization/deserialization across Internet domain sockets first. :-)
View project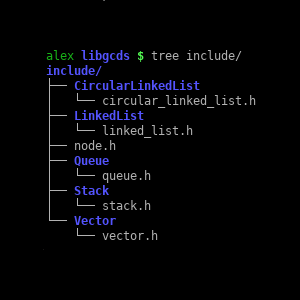
libgcds (personal project)
A library for generic C data structures such as stacks, queues, linked lists, or vectors. Although libgcds may be compiled from source, it is also distributed as a static library that you can use by simply linking it to your projects using -lgcds with gcc.
View project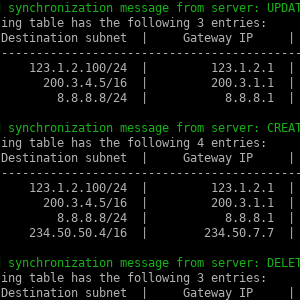
Linux-IPC (personal project)
A collection of Linux inter-process communication (IPC) mechanisms implemented in C: UNIX domain sockets, message queues, shared memory, and signals (TODO). These IPC mechanisms are jointly applied for simulating routing and ARP table managers running on a server and being synchronized across every connected client process.
View project
CoMetGeNe (PhD project)
A bioinformatics tool that I developed in Python during my PhD. It identifies longest sequences of reactions in the metabolic pathways of a query organism such that the genes involved in these reactions are neighbors on the chromosome. No manual data download is necessary, as CoMetGeNe retrieves the required data automatically from the KEGG database for species of your choosing. It is also possible to study conserved neighborhood patterns in metabolic and genomic contexts for several species. In order to facilitate the analysis of multi-species data sets, CoMetGeNe takes full advantage of multiprocessing.
View project
Genome coverage (Master's project)
A bioinformatics tool that takes as input genomes and genomic sequences called "reads", then computes and displays genome coverage curves. Genome coverage at a given position translates to the number of reads that overlap that particular position. The problem of (exact) string matching is handled using an optimized version of suffix arrays, thus rendering it very fast in practice. GenomeCoverage is written in Java, the GUI is built with Swing, and genome coverage graphics are generated with JFreeChart.
View project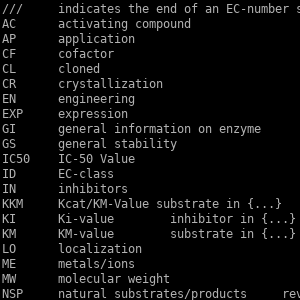
BRENDA-Parser (personal project)
BRENDA is an extremely valuable resource for studying metabolism as it is the knowledge base that contains the most detailed information on enzymatic activities. Unfortunately it is not free, not even for academics. They do offer a free version though, which comes as a flat file with notoriously horrendous formatting. The formatting is so bad I haven't been able to find a parser that actually parses the flat file correctly, so I wrote BRENDA-Parser in Python for this purpose.
View project